Literature Sharing: Exploring STING Expression Characteristics in Colorectal Cancer Using Multiplex Fluorescence
1. Research Background
Colorectal cancer (CRC) is one of the most common cancers globally and the second leading cause of cancer-related deaths. Current standard treatment options have limited efficacy, especially for advanced metastatic patients, with a 5-year survival rate of only 13%. Immunotherapy such as immune checkpoint inhibitors is effective in only about 15% of patients with mismatch repair deficiency or microsatellite instability (MSI) subtypes, thus there is an urgent need to explore new therapeutic targets.
The stimulator of interferon genes (STING) holds promise as a potential anti-cancer therapeutic target. As a nucleic acid-sensing pattern recognition receptor, STING can detect DNA damage and pathogen infection, playing a bridging role in the type I interferon signaling pathway, and is a key molecule in innate and anti-tumor immune activation. STING agonists can initiate type I interferon-dependent responses in tumor cells and immune cells, and when used alone or in combination with existing anti-cancer drugs, they are expected to produce anti-tumor effects mediated by immune cells.
STING exhibits multi-cell type expression characteristics in cancer tissues, including lymphocytes, natural killer cells, endothelial cells, and epithelial cells. Current studies on STING expression in the cancer field are mostly based on RNA levels, which makes it difficult to distinguish cell-specific expression differences, and the clinical significance of STING in tumor cells and tumor-associated inflammatory cells is not yet fully clear. Based on this, a research paper published in Pathology titled "High-level STING expression in tumour and inflammatory cells is linked to microsatellite instability and favourable tumour parameters in a cohort of over 1,900 colorectal cancer patients" used multiplex fluorescent immunohistochemistry (mIHC) technology to quantitatively analyze STING expression in different cell types in colorectal cancer and explore its association with clinicopathological characteristics. This provides important basis for the study of immune mechanisms and potential therapeutic target exploration in colorectal cancer.
2. Research Methods
The study collected tissue samples from clinically confirmed colorectal cancer patients and constructed a tissue microarray (TMA) containing 2418 colorectal cancer tissue samples. To ensure sample quality, all tissue samples were re-examined and confirmed by pathologists, and samples with incomplete tissue morphology or unclear diagnosis were excluded, resulting in 1905 interpretable samples. Multiplex fluorescent immunohistochemistry technology combined with deep learning image analysis algorithms was used to achieve precise quantitative detection of STING expression in specific cell types. The experimental process included: deparaffinization to water of tissue microarrays, antigen retrieval (high-temperature repair with citrate buffer), blocking of non-specific binding sites, addition of STING primary antibody and specific fluorescently labeled secondary antibody, and acquisition of images through a high-resolution scanner after fluorescent color development. Deep learning algorithms were used to segment and analyze the images, automatically identify different cell types, and quantitatively count the number of STING-positive cells, relative expression intensity, and proportion of positive cells in each cell type. To verify the reliability of the detection results, traditional bright-field IHC technology was used for parallel detection of some samples, staining with STING-specific antibodies, and evaluation of expression levels through blind reading by pathologists to ensure the consistency of results between the two detection methods. Finally, R software and JMP Pro 17 statistical software were used for data processing and analysis.
3. Core Results
Multiplex Fluorescent Immunohistochemistry Detection of STING in Tumor Cells
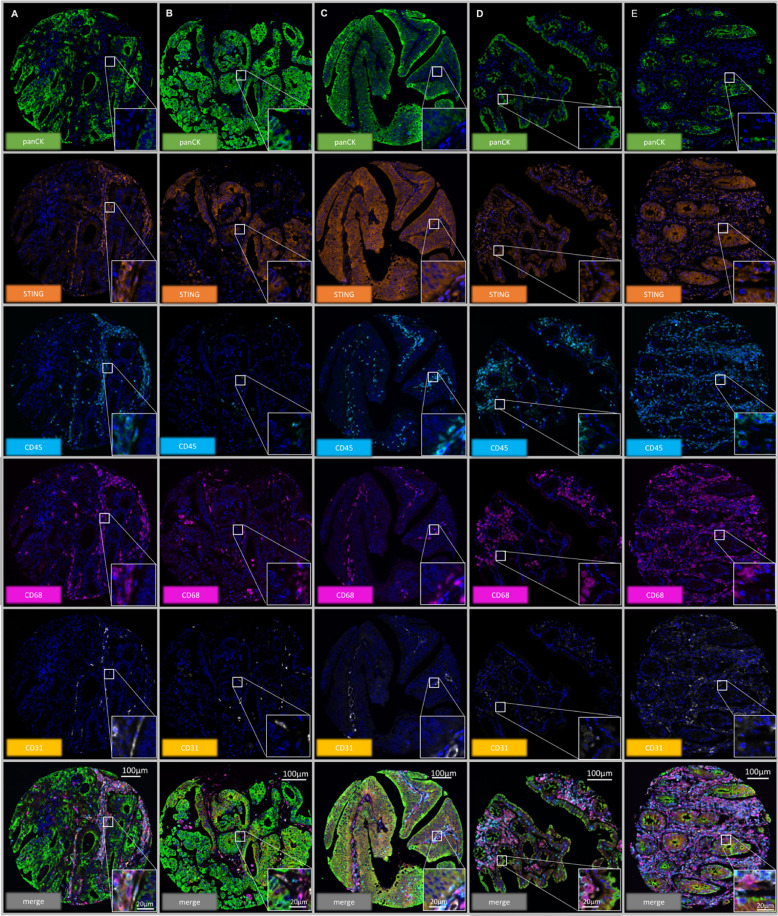
The above figure mainly shows the cellular localization and expression pattern of STING protein in colorectal cancer tissues (A-C). The results show that STING is expressed to varying degrees in tumor cells, macrophages, CD68- leukocytes, and endothelial cells, and its expression in tumor cells is highly heterogeneous, ranging from completely negative to strongly positive. At the same time, the figure also presents the differences in STING staining intensity and distribution between normal colonic epithelium (D, E) and cancer tissues, intuitively confirming the widespread existence of STING in the tumor microenvironment and the complexity of its expression, providing important morphological basis for subsequent analysis of the association between STING and clinicopathological characteristics such as MSI, tumor stage, and immune infiltration.
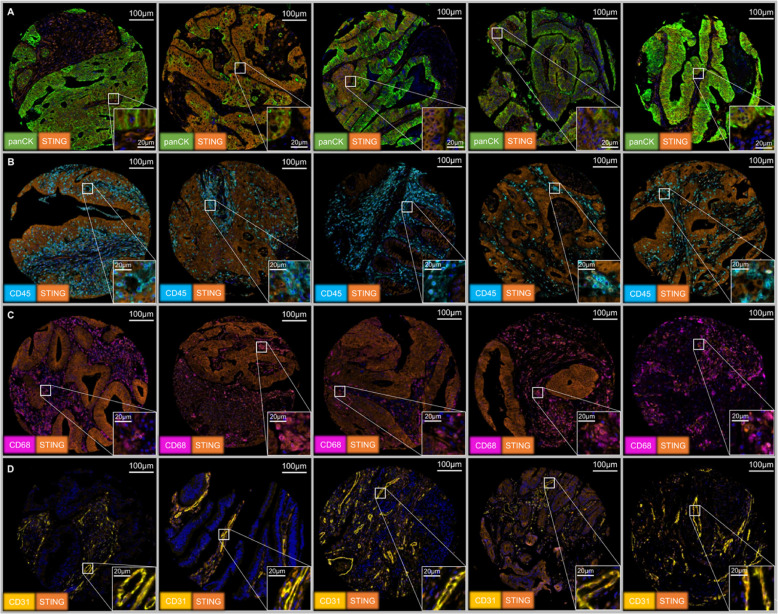
This figure further shows the immunohistochemical expression characteristics of STING in colorectal cancer tissues, focusing on its staining patterns in different cell types and heterogeneous distribution in tumor tissues. It includes representative images from different cases, showing that STING can be negative, weakly positive, or strongly positive in tumor cells, while also showing obvious staining in macrophages, lymphocytes, and endothelial cells in the tumor stroma. Through these images, the study intuitively confirmed the widespread expression of STING in the tumor microenvironment and the high variability of its expression levels.
Multi-dimensional Analysis of STING Expression in Colorectal Cancer
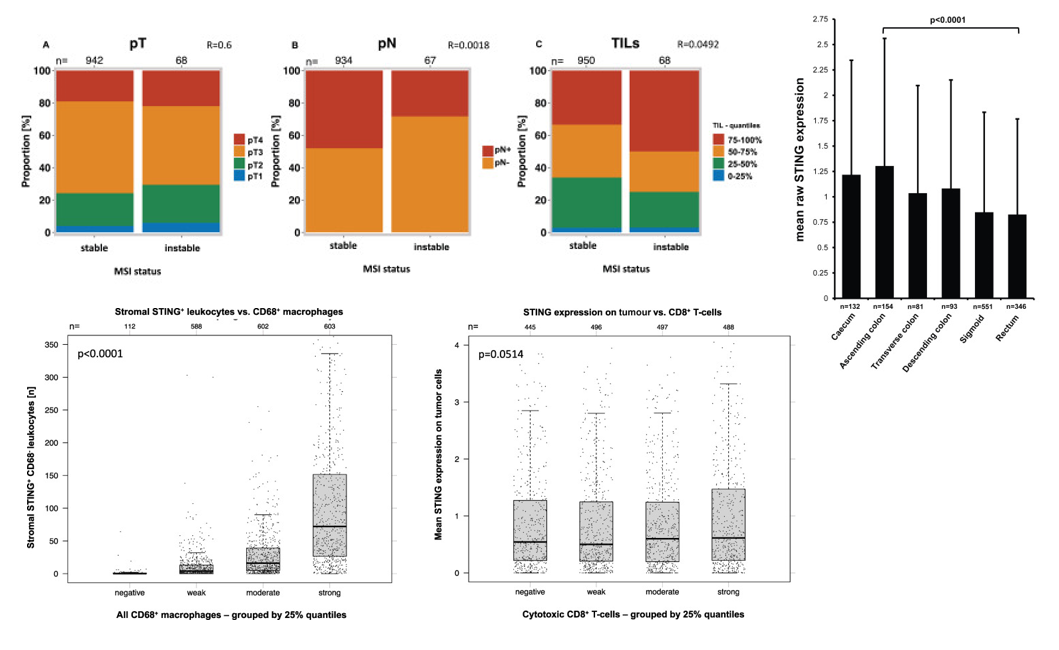
The study further revealed the complex association between STING expression and tumor progression and immune microenvironment through series analysis: Stacked bar charts first clarified that MSI/MSS status is independent of tumor invasion depth (pT) and only weakly associated with lymph node status and immune cell count, indicating that molecular subtype is not a key factor determining tumor invasion depth. On this basis, it further showed significant site differences in STING expression in tumor cells, with the highest expression in the ascending colon and the lowest in the rectum, suggesting that anatomical location may shape tumor biological behavior by affecting STING pathway activity. From the perspective of immune cell interaction, it was found that high infiltration of STING-positive CD68-negative leukocytes in the stroma was significantly correlated with the total number of macrophages, indicating that the enrichment of STING-positive lymphocytes is often accompanied by enhanced overall immune infiltration; finally, comparison with previous studies showed that although STING expression in tumor cells showed a slight upward trend with the increase in CD3+ and CD8+ cytotoxic T cell counts, it did not reach statistical significance, indicating that T cell infiltration has limited impact on the intrinsic STING pathway in tumor cells.
These results collectively indicate that STING expression is regulated by multiple factors including anatomical location and immune microenvironment, and its different change patterns in tumor cells and immune cells may synergistically participate in the progression of colorectal cancer.
STING Expression Analysis in Bright-field Immunohistochemistry
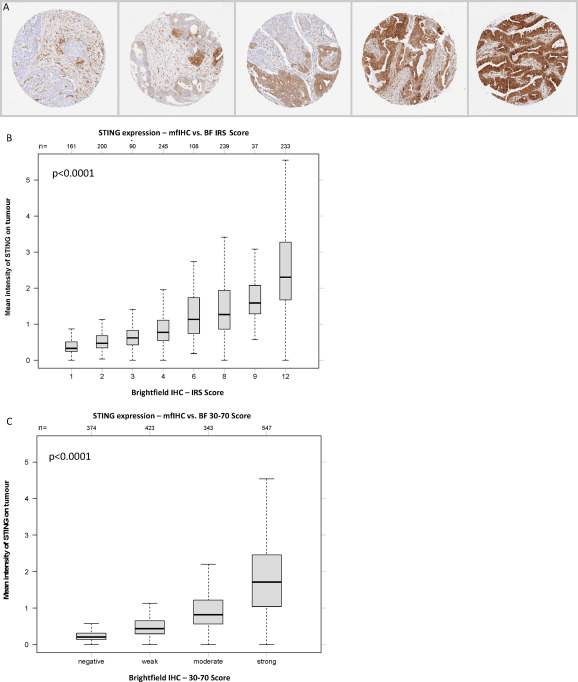
To ensure the reliability and accuracy of multiplex fluorescent immunohistochemistry detection results, the study designed strict parallel validation experiments. Representative colorectal cancer samples were selected, and both traditional bright-field IHC technology and mIHC technology were used to detect STING expression levels in tumor cells. Among them, bright-field IHC used DAB as a chromogen and was scored based on the proportion of positive cells and staining intensity. mIHC obtained STING expression data through quantitative analysis of fluorescent signals. The study limited STING staining assessment to tumor cells and found significant differences in expression between different tumors, with a positive rate of 77.7% and mainly strong positivity, while bright-field IHC and mfIHC showed highly consistent results for high STING expression in tumor cells.
4. Summary
Through rigorous experimental design and large-sample analysis, this study systematically clarified the expression characteristics and clinical associations of STING in different cell types of colorectal cancer, providing important clinical basis for the study of immune mechanisms and development of therapeutic targets in colorectal cancer. In the future, further exploration of the molecular mechanisms by which STING regulates the tumor immune microenvironment and clinical studies of STING agonists combined with other treatment regimens can be conducted based on the results of this study.
References
Bady E, Ebner J, Mandelkow T, Huang Z, Neipp M, Mofid H, Lárusson H, Daniels T, Isbert C, Coerper S, Ditterich D, Rupprecht H, Goetz A, Bernreuther C, Sauter G, Uhlig R, Wilczak W, Simon R, Steurer S, Minner S, Burandt E, Krech T, Hackert T, Melling N, Schraps N, Clauditz TS, Marx AH, Müller JH. High-level STING expression in tumour and inflammatory cells is linked to microsatellite instability and favourable tumour parameters in a cohort of over 1,900 colorectal cancer patients. Pathology. 2025 Dec;57(7):843-854. doi: 10.1016/j.pathol.2025.05.008. Epub 2025 Jul 14. PMID: 40816937.
Enkilife mIHC TSA Kits
Product | Catalog Number |
|---|---|
TSA Six-Label Seven-Color Multiplex Immunohistochemistry Kit | |
TSA Five-Label Six-Color Multiplex Immunohistochemistry Kit | |
TSA Four-Label Five-Color Multiplex Immunohistochemistry Kit | |
TSA Three-Label Four-Color Multiplex Immunohistochemistry Kit | |
TSA Two-Label Three-Color Multiplex Immunohistochemistry Kit |
