TSA-mIF for TIME Analysis
Background
The interaction between cancer and the host immune system mainly occurs in the reactive tissue stroma surrounding tumors, an area known as the Tumor Immune Microenvironment (TIME). The breakthrough clinical application of immune checkpoint inhibitors, particularly targeted therapies against Programmed Death Protein 1 (PD-1, a T-cell co-inhibitory receptor) and Programmed Death Ligand 1 (PD-L1, also known as B7-H1/CD274), has not only propelled cancer immunotherapy into a new era but also made functional analysis of TIME a core focus in both research and clinical settings. Precise characterization of TIME's cellular composition, functional states, and molecular interaction networks is a crucial prerequisite for screening predictive biomarkers for immunotherapy response. Furthermore, deeply clarifying the dual pro-cancer/anti-cancer mechanisms of various immune cells in TIME and their dynamic associations with tumor cells can provide core support for developing novel immunotherapeutic strategies. However, the high heterogeneity of TIME and the challenge of detecting low-abundance immune molecules pose severe challenges to traditional analytical techniques, urgently requiring highly sensitive, multiplex-targeted in situ detection tools to overcome this dilemma.
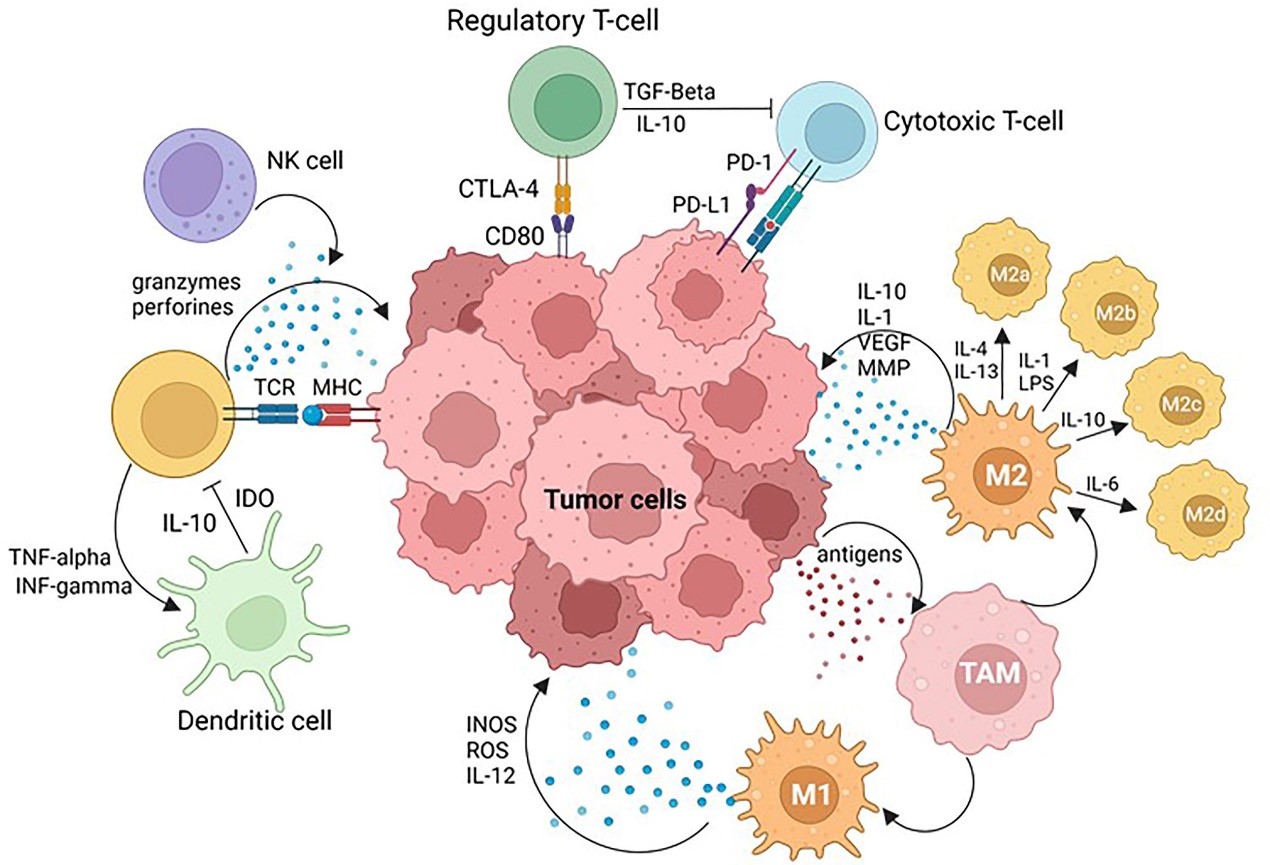
Tumor Microenvironment (TME)[1]
Advantages
For over a century, hematoxylin-eosin staining (HE staining) has been the classic method for pathologists to observe lymphocytes, cancer cell nuclei, glands, and other tissue structures in biopsy and surgical samples under microscopic examination. Its morphological characteristics provide key evidence for disease grading and diagnosis. Immunohistochemistry (IHC) is a routine method for analyzing clinical markers such as estrogen receptors and human epidermal growth factor receptor 2, but both have obvious limitations in TIME research — HE staining cannot accurately identify immune cell subsets and functional molecules, and standard IHC can only detect one marker per section, making it difficult to capture the complex characteristics of multicellular interactions and molecular networks in TIME. In recent years, multiplex immunofluorescence technology (mIF) based on the principle of Tyramide Signal Amplification (TSA) has completely overcome this dilemma. Through the core strategy of "single marker antibody labeling - horseradish peroxidase (HRP) catalyzing covalent binding of fluorophore-conjugated tyramide molecules to target epitopes - antibody elution for multiple rounds of cyclic staining", combined with multispectral imaging microscopy scanning and AI image analysis, this technology can simultaneously detect 5-10 markers on a single tissue section. It not only can accurately identify immune subsets such as CD8⁺T cells, regulatory T cells, and tumor-associated macrophages, and achieve highly sensitive detection of low-abundance immune checkpoint molecules (such as PD-1/PD-L1), but also can completely preserve the in situ tissue structure, quantify the spatial proximity relationship and interaction patterns between cells. It solves the problem that traditional technologies are difficult to simultaneously obtain multidimensional information of "cell typing - functional phenotype - spatial distribution", and provides strong technical support for analyzing the immunosuppressive mechanisms of TIME, screening biomarkers for immunotherapy response, and discovering key functional regions such as tertiary lymphoid structures (TLS), becoming a core tool for tumor immunology research and precision diagnosis and treatment transformation.
Cases
Using TSA-based multiplex immunofluorescence technology, Maltseva et al. systematically analyzed the TIME immune cell composition and PD-1 expression characteristics in patients with immunotherapy response and non-response groups. This study achieved precise identification and classification of cells through inForm phenotypic analysis software, ensuring the objectivity and efficiency of data analysis. The experiment selected a targeted antibody panel including CD8, PD-1, CD20, CD163, and FoxP3 antibodies, targeting cytotoxic T lymphocytes, immune checkpoint molecules, B lymphocytes, pro-tumor phenotypic macrophages, and regulatory T cells, respectively. Nuclei were stained with DAPI and treated with anti-fluorescence quenching mounting medium to maintain signal stability. During the quantitative analysis phase, 10 representative fields of view were selected under 200x magnification, and the percentage of the above immune cells in the total stromal cells within tumor nodules (excluding the surrounding stroma of tumor cells and tertiary lymphoid structures) was calculated. Through multi-color labeling (CD8⁺ cells green, CD20⁺ cells yellow, CD163⁺ cells red, Treg cells orange, PD-1 expression white, nuclei blue), the cell infiltration pattern was visually presented. Meanwhile, InForm software was used to convert single marker data into immunohistochemical simulation maps to further assist in result interpretation. Through the high sensitivity and multiplex detection advantages of TSA technology, this study clearly revealed the differences in TIME immune landscape between patients with different treatment response groups, providing direct experimental evidence for the screening of immunotherapy response biomarkers, and also confirming the core application value of TSA multiplex immunofluorescence technology in the in-depth analysis of TIME in clinical samples.
![]()
![]()
![]()
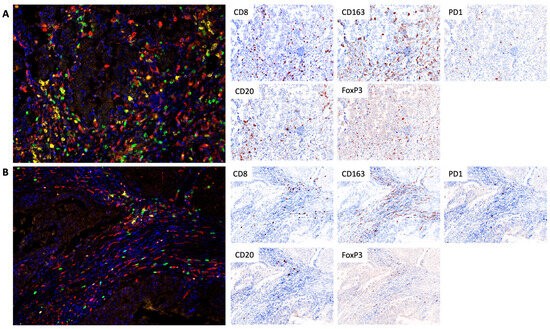
Figure 1 Tumor infiltration of CD8 cytotoxic lymphocytes, CD20 B lymphocytes, FoxP3 regulatory T cells, and CD163 macrophages in responders (A) and non-responders (B)[2].
In a study of the tumor immune microenvironment in non-small cell lung cancer (NSCLC), Peng et al. systematically analyzed the expression characteristics and interaction relationships of 10 immune markers in tumor nests (TN) and tumor stroma (TS) using multiplex immunofluorescence technology. The results showed multiple strong correlative immune cell interactions within the TIME: stromal CD20+ B cells with stromal CD4⁺CD38⁺T cells, neutrophils with FOXP3+ cells, stromal neutrophils with intratumoral regulatory T cells, intratumoral CD38+ T cells with intratumoral CD20+ B cells, intratumoral CD8+ T cells with intratumoral PD-L1-expressing M2 macrophages, and intratumoral PD-L1+ cells with intratumoral CD8+ T cells. Additionally, the study found moderate correlations between intratumoral CD133+ cells and intratumoral M1 macrophages, particularly M1 macrophages not expressing PD-L1. This suggests that intratumoral macrophages may play a role in mediating CD8⁺T cell exhaustion-type immune responses. Through the multiplex detection advantages of multiplex immunofluorescence technology, this study clearly outlined the TIME immune landscape and cell interaction network of NSCLC, providing important references for subsequent mechanism exploration and therapeutic target screening.

Figure 2 Expression profiles of 10 immune markers in non-small cell lung cancer. The figure shows individual and merged immunofluorescence images, as well as pathological sections[3].
In a tissue microarray (TMA) study, Mori et al. stained bronchial airway tumor and breast cancer TMA samples using optimized multiplex immunofluorescence protocols (IP1 and IP2). Multi-spectral image unmixing and analysis were completed using inForm software, while generating "pathological views" simulating DAB staining to assist in result interpretation. Among them, the IP1 protocol can simultaneously identify 5 cell types in the TME (CD3⁺T cells, CD20⁺B cells, CD3⁺FOXP3⁺regulatory T cells, CD117⁺CK⁻mast cells, CK⁺epithelial/tumor cells). Combined with Ki67 staining, cell proliferation status can be evaluated, and different cell subsets can be distinguished through co-localization of CD117 and CK. The IP2 protocol focuses on 4 core cell types (CD3⁺CD8⁻T cells, CD3⁺CD8⁺cytotoxic T cells, CD68⁺macrophages, CK⁺epithelial/tumor cells) and incorporates PD-1/PD-L1 immune checkpoint molecule detection, successfully capturing the co-localization characteristics of PD-L1⁺CD68⁺macrophages and PD-1⁺T cells in breast cancer samples. Through the combination of standardized mIF protocols and TMA technology, this study efficiently achieved systematic analysis of TME cell composition, functional status, and immune molecule interactions in multiple types of tumor samples, providing a feasible path for high-throughput clinical sample TIME research.
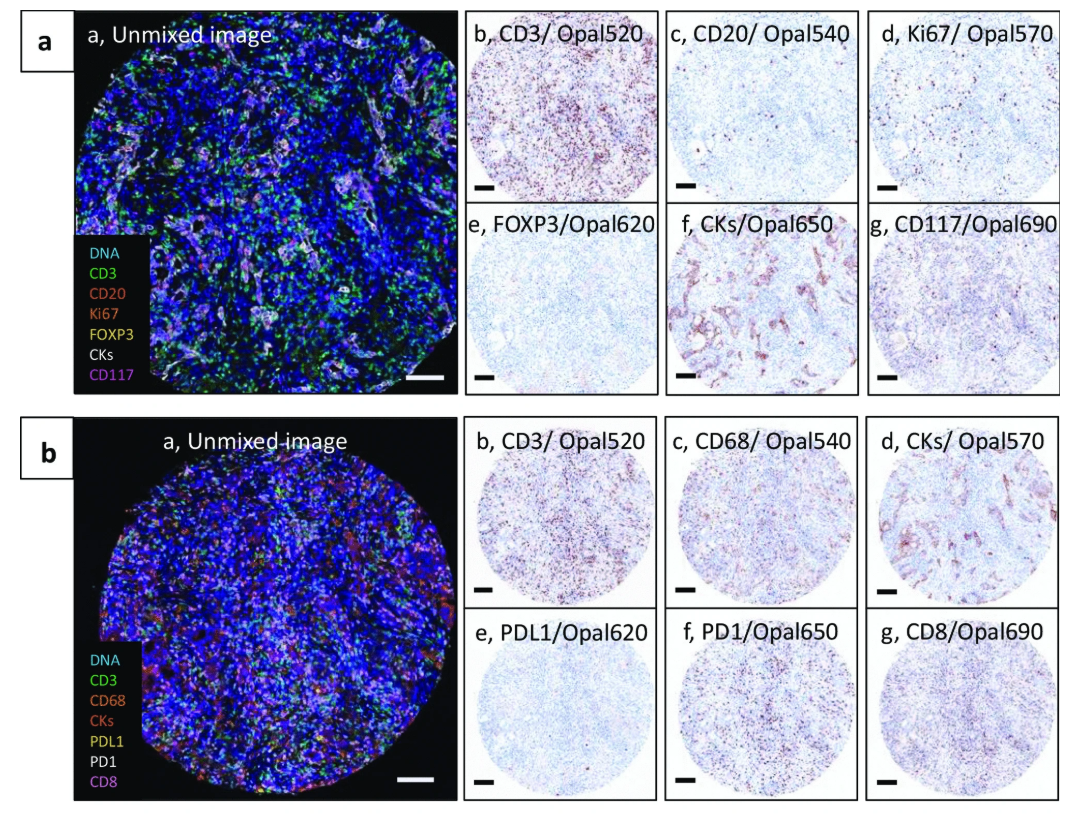
Figure 3 Human bronchial airway biopsy tissue microarrays stained using two multiplex immunofluorescence protocols, IP1 (a) and IP2 (b)[4].
Enkilife can provide customers with TSA immunohistochemistry and immunofluorescence section analysis services, including positive cell count analysis, co-localization analysis, proximity analysis, cell density analysis, tumor cell infiltration analysis, etc.
Product | Catalog Number |
|---|---|
TSA Six-Label Seven-Color Multiplex Immunohistochemistry Kit | |
TSA Five-Label Six-Color Multiplex Immunohistochemistry Kit | |
TSA Four-Label Five-Color Multiplex Immunohistochemistry Kit | |
TSA Three-Label Four-Color Multiplex Immunohistochemistry Kit | |
TSA Two-Label Three-Color Multiplex Immunohistochemistry Kit |
For details, please check TSA mIHC Kit
References
1. Rojas F, Hernandez S, Lazcano R, Laberiano-Fernandez C, Parra ER. Multiplex Immunofluorescence and the Digital Image Analysis Workflow for Evaluation of the Tumor Immune Environment in Translational Research. Front Oncol. 2022 Jun 27;12:889886. doi: 10.3389/fonc.2022.889886. PMID: 35832550; PMCID: PMC9271766.
2. Maltseva A, Kalinchuk A, Chernorubashkina N, Sisakyan V, Lots I, Gofman A, Anzhiganova Y, Martynova E, Zukov R, Aleksandrova E, Kolomiets L, Tashireva L. Predicting Response to Immunotargeted Therapy in Endometrial Cancer via Tumor Immune Microenvironment: A Multicenter, Observational Study. Int J Mol Sci. 2024 Apr 1;25(7):3933. doi: 10.3390/ijms25073933. PMID: 38612743; PMCID: PMC11011874.
3. Peng H, Wu X, Zhong R, Yu T, Cai X, Liu J, Wen Y, Ao Y, Chen J, Li Y, He M, Li C, Zheng H, Chen Y, Pan Z, He J, Liang W. Profiling Tumor Immune Microenvironment of Non-Small Cell Lung Cancer Using Multiplex Immunofluorescence. Front Immunol. 2021 Nov 4;12:750046. doi: 10.3389/fimmu.2021.750046. PMID: 34804034; PMCID: PMC8600321.
4. Mori H, Bolen J, Schuetter L, Massion P, Hoyt CC, VandenBerg S, Esserman L, Borowsky AD, Campbell MJ. Characterizing the Tumor Immune Microenvironment with Tyramide-Based Multiplex Immunofluorescence. J Mammary Gland Biol Neoplasia. 2020 Dec;25(4):417-432. doi: 10.1007/s10911-021-09479-2. Epub 2021 Feb 15. PMID: 33590360; PMCID: PMC7960613.
